A Scientific Journey with Professor Jonathan A. King to Uncover Bacteriophage Assembly Process by Pushing the Boundaries of Cryogenic Electron Microscopy and Tomography
Wah Chiu
Wallenberg-Bienenstock Professor
Stanford University, Stanford, California
wahc@stanford.edu
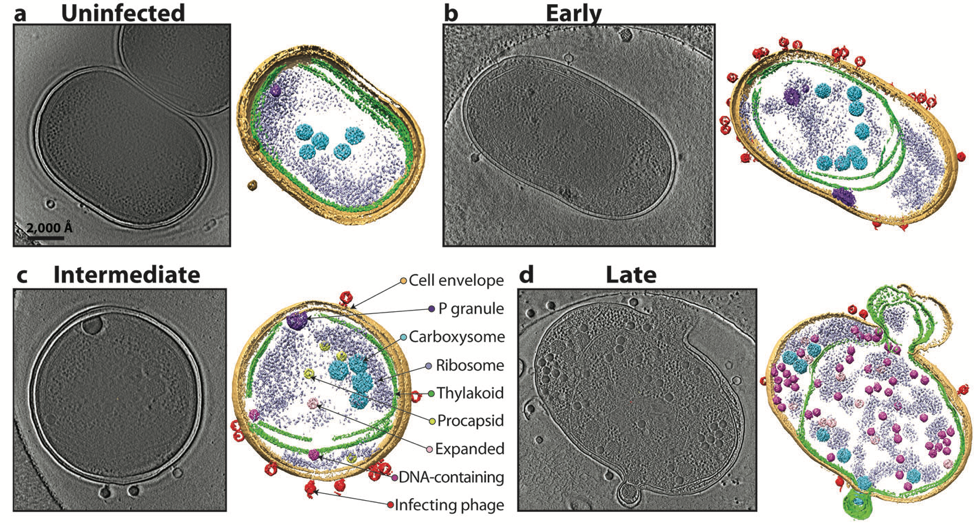
For nearly 40 years, I have known Jonathan King through our collaboration on cryogenic electron microscopy (cryoEM) of bacteriophages. In the late 1980s, single particle cryoEM was in its infancy and I thought virus particles would be good biological assemblies to gain insights into virus assembly principles with the use of cryoEM. Peter Prevelige, then a postdoc of Jon, introduced me to studying structures of P22 bacteriophage. In 1993, our first manuscript in Journal of Molecular Biology described the use of cryoEM to resolve the structure differences of the P22 phage in different morphological states at ~40 Å resolution for the first time [1]. Since then, King’s lab has been generously and enthusiastically supporting our research to pursue high resolution structure determination of bacteriophages. We finally obtained a near atomic resolution (~4.5 Å) cryoEM structure for the epsilon15 phage (Figure 1) in 2008. Our Nature paper showed for the first time not only the de novo c-alpha atom backbone model of the major capsid protein but also an identification of a previously unknown small capsid protein in this virion [2]. This groundbreaking work was the first near atomic resolution single particle cryoEM structure determination without any prior crystal structure information of its component prior to the “resolution revolution” era of cryoEM. Moreover, both our labs also pioneered the use of cryogenic electron tomography and subtomogram averaging to identify multiple assembly intermediates of bacteriophages in a cyanobacterium using a new electron optics called Zernike phase plate for image contrast enhancement in a 200 kV electron microscope (Figure 2) which was reported in Nature in 2014 [3].
I am very grateful to Jonathan King and his lab members including Peter Prevelige, Peter Weigele, and Cameron Haase-Pettingell who have worked tirelessly alongside my lab members studying the bacteriophage structures both in vitro and in cellulo for many years. Our collaboration has facilitated my research effort in pushing the boundaries of single particle cryoEM structure determination from nanometer to near atomic resolution and his patience and support made it not only possible, but also a very enjoyable scientific journey.
In addition to our cryoEM partnership, Jon and I collaborated on another significant research adventure in 2007. We initiated a Nanomedicine Center supported by NIH Roadmap grant with 10 other investigators around the country aimed at engineering new protein folding machines with new functionalities. Jon was a very insightful and active participant whose creative ideas and immense knowledge in protein folding and homeostasis in relation to human diseases benefited our team projects [4]. For over a decade, we made many exciting progresses towards a very bold project goal and established many follow-up collaborations among our team members from Baylor College of Medicine, M.I.T., Stanford, MD Anderson Cancer Center, UC San Diego, UC Irvine and UCSF.
In total, Jonathan King and I had 24 joint publications with many exciting discoveries at the time. On a personal level, Jonathan is a very generous, honest and open individual. I visited his lab regularly in Cambridge throughout these years. I had many memorable times including a short break with his wife, Jackie and their friends in the South Shore in one summer.
I wish Jonathan and his family well, happy and healthy in the years to come.

References
[1]. Prasad BV, Prevelige PE, Marietta E, Chen RO, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol 231:1, 65-74 (1993).
[2]. Jiang W, Baker ML, Jakana J, Weigele PR, King J, Chiu W. Backbone structure of the infectious epsilon15 virus capsid revealed by electron cryomicroscopy. Nature 451:7182,1130-4 (2008).
[3]. Dai W, Fu C, Raytcheva D, Flanagan J, Khant HA, Liu X, Rochat RH, Haase-Pettingell C, Piret J, Ludtke SJ, Nagayama K, Schmid MF, King JA, Chiu W. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature 502:7473, 707-10 (2013).
[4]. Sergeeva OA, Chen B, Haase-Pettingell C, Ludtke SJ, Chiu W, King JA. Human CCT4 and CCT5 Chaperonin Subunits Expressed in Escherichia coli Form Biologically Active Homo-oligomers. J Biol Chem 288:24, 17734-44 (2013).

