Amino Acid Side Chains Directing Tailspike Folding, Assembly and Maturation
Scott Betts
scottbetts1@gmail.com
I first met Jon King in 1995 after I invited him to travel to The University of Michigan to talk about his P22 tailspike research. At the time I was a graduate student in the NIH Cellular Biotechnology training program and at a meeting of fellow trainees, a request was made for a volunteer to organize the annual symposium sponsored by the program. This involved inviting external speakers to present their research on some topic relevant to the program. I broke a long, uncomfortable silence by offering to organize the symposium – on the condition that I could choose the topic as well as the speakers. To my surprise, my busy fellow trainees gladly and unanimously accepted my offer.
I knew exactly who I wanted to invite to the symposium – MIT Professor Jonathan King. I had spent my first couple of years as a graduate student making point mutations in the psbO gene of spinach photosystem II, expressing these mutant genes in E. coli, refolding them from inclusion bodies, and then mixing the soluble recombinant proteins with isolated photosystem II complexes in vitro. The objective of these experiments was to understand how the psbO gene product, the Manganese-Stabilizing Protein, activates water-splitting activity in the oxygen-evolving complex of photosystem II. During this time the title of an article in Science caught my eye – Global suppression of protein folding defects and inclusion body formation. In my entire career I have never read an article with such delight as when I first read this one by Anna Mitraki et al. It was mind blowing.
Of course, Jon graciously accepted my invitation extended somewhat fraudulently “on behalf of my fellow NIH trainees” to speak at the symposium on my chosen topic of protein folding in vivo. And sorry Jon for the invitation under false pretenses – but I have no regrets of course! I hosted Jon’s visit, and that of Stanford professor Robert Baldwin, who I invited for some ideological balance (insert wink emoji), to Ann Arbor. As I recall by the end of the day of the symposium, I had coerced an invitation to visit Cambridge and present my dissertation research to Jon’s group. This eventually led to a postdoc appointment in the NSF Biotechnology Process Engineering Center (BPEC) at MIT and in Jon’s lab in the Department of Biology. It was honestly a dream come true for the youngest child of two labor union members from Milwaukie, Oregon, and with the first PhD in my extended family.
When I started working in Jonathan’s lab, I spent a lot of time in the BPEC lab located in Building 10. There was a nice Pharmacia FPLC chromatography instrument there that I used to purify tailspike protein. I remember lamenting once to lab mates that we did not have an FPLC in Jon’s lab, and I think it was one of the Chem E grad students who mentioned the existence of another, unused FPLC that was stored in a closet in Building 10. I couldn’t believe my luck that such an expensive instrument might be available as surplus equipment and was delighted to locate it and get permission to dust it off and set it up in Jon’s lab where it performed beautifully. I thought I had finally found the promised land where expensive equipment was sometimes forgotten in closets.
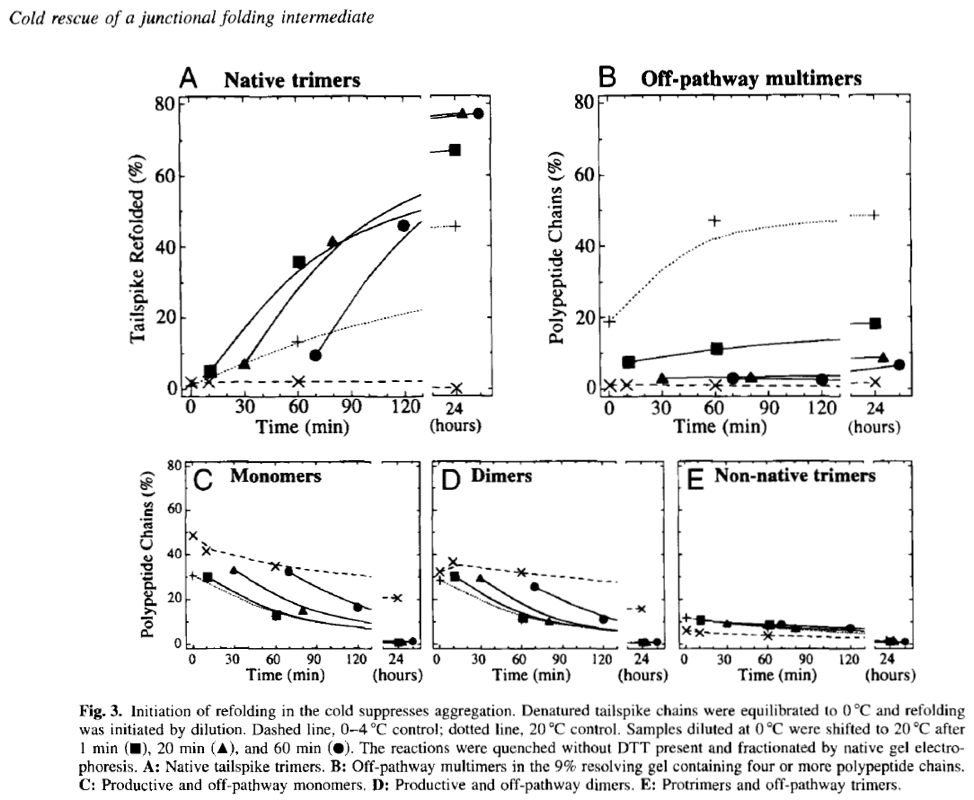
As was the case for many King lab newbies, I spent many of my early days in the lab learning from Cammie how to grow Salmonella cultures and infect them with P22 phage selected from the lab’s vast collection of mutants. The cultures were the production system for P22 tailspike protein for our experiments. We purified, denatured, and refolded various mutant tailspikes under different conditions, attempting to trap intermediates on both the folding and aggregation pathways and then fractionate them using native gel electrophoresis. An exciting early finding came from an experiment where we initiated refolding of denatured tailspikes on ice, and then shifted the reactions to warmer temperatures. We found that by initiating refolding at low temperatures we could trap and accumulate single tailspike chains in an assembly-competent conformation and “rescue” them from the non-productive aggregation pathway that Margaret Speed and Jonathan had characterized prior to my arrival in the lab. It was amazing to see these monomeric and productive folding intermediates accumulate and be resolved and detectable on native polyacrylamide gels. This further enabled quantitation of the various folding intermediates as they accumulated and disappeared over a time course of hours (see Fig. 3 below from (Betts and King 1998)).
As part of some more original experiments that I initiated later in my King lab tenure, Jon connected me with Patricia Reilly in the electron microscopy facility and asked her to teach me how to prepare micrographs of E. coli cells. We were hoping to visualize inclusion bodies formed from over-expressed mutant tailspike proteins. I assisted Patricia in the fixation and sectioning of cells and she generated some amazing images of individual E. coli cells with heavily stained inclusion bodies at one or both poles (see micrograph in pathway diagram below from (Betts and King 1999) and in the unpublished images below that). As an aside, this same diagram shows an “inclusion body precursor” polypeptide drawn by my hand and which ends at the C-terminus (or at least the right end in the drawing) with the cursive letters Max. This cryptic scribble was intended as a little time capsule for my son, Max, who at that time was a first grader at Somerville Elementary School.
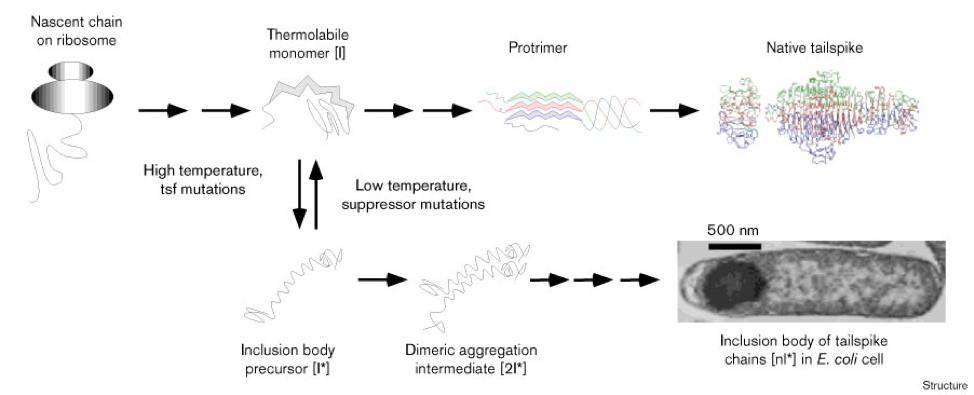
Figure 3. Competing folding and inclusion body pathways. The single-chain intermediate [I] is the junctional thermolabile intermediate at the branch-point between the productive and inclusion body pathways. The protrimer has interchain disulfide bonds [5]. For further details see text.
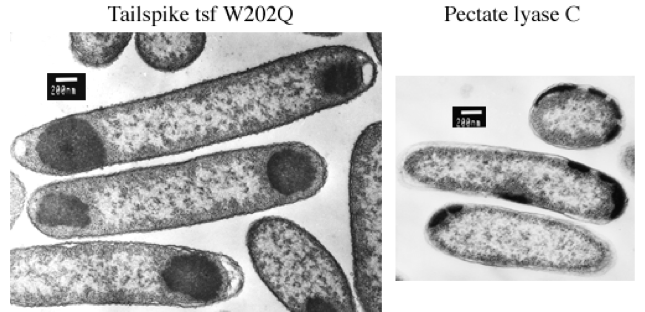
Left: During synthesis at 37°C, tailspike polypeptides containing tsf mutations self-associate in the cytoplasm of host E. coli cells. The aggregated polypeptide chains accumulate as electron-dense inclusion bodies visible at one or both poles of the cells in these electron micrographs.
Right: In E. coli, recombinant pectate lyase C is targeted to the periplasmic space by its native signal peptide, where at low temperature it accumulates in the processed, mature form. At 37°C, however, pectate lyase C aggregates and accumulates as electron-dense inclusion bodies that appear to be excluded from the cytoplasm. The location of pectate lyase C inclusion bodies at the cell periphery suggests that they form in the periplasmic space. Sample processing and EM by Patricia Reilly at the Biomedical Electron Microscopy facility at MIT. The plasmid encoding the pectate lyase C precursor was kindly provided by Noel Keen and Frances Jurnak.
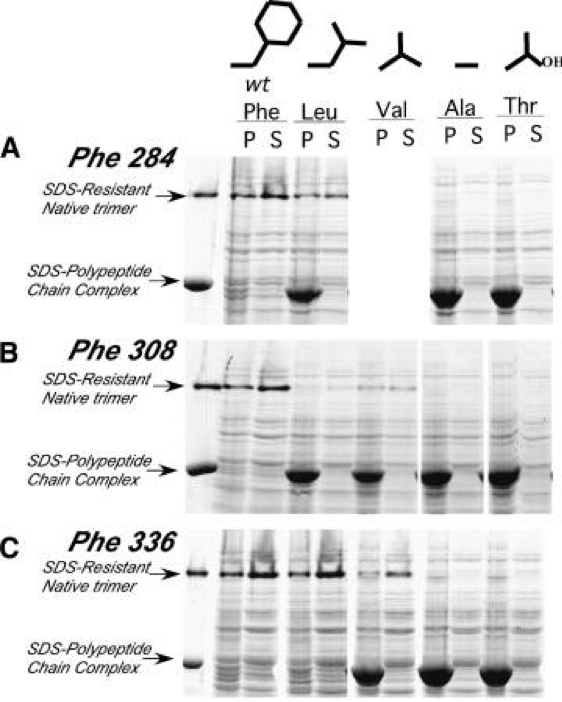
The problem that Jon and I decided to focus on for the remainder of my postdoctoral training was the role of buried amino acid side chains in the folding of the parallel beta-helix domain in the P22 tailspike. As shown in the diagram below (Fig 1 in (Betts, Haase-Pettingell et al. 2004)) buried amino acid side chains (cyan and green shading) in this structure line up in neat stacks or rungs. Temperature sensitive folding mutations had never been recovered in these buried stacks using screens that required recovery of viable phage. This observation suggested that these buried side chain stacks were absolutely required for tailspike folding. We decided to attempt to demonstrate this experimentally by expressing subunits with amino acid replacements in buried stacks in E. coli, probably as inclusion bodies, and then attempting to refold them in vitro into tailspike trimers.
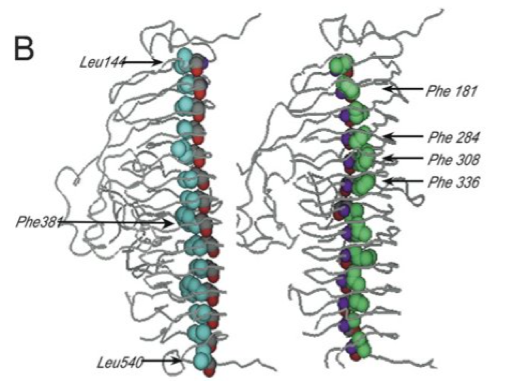
Figure 1B (Betts, Haase-Pettingell et al. 2004). Depiction of the single parallel β-helix domain showing two of the three interior side-chain stacks, stack A (cyan) and stack C (green); the backbone atoms are shown using CPK color scheme residues mutated in this paper are indicated.
We generated a series of mutant tailspike genes encoding increasingly smaller hydrophobic side chains and a polar side chain at each of the side chains labeled in the above figure. These mutations of course were all generated in the lab using the QuikChange mutagenesis kit from Stratagene, a real breakthrough at a time when researchers did their own mutagenesis. Nowadays it is more efficient and cost effective to pay a third party to synthesize and clone DNA containing the mutations, but such services were not available to 20th Century molecular biologists. Also, during my time in the King lab (1996-1999) I poured my last DNA sequencing gel and we started “sequencing by mail” sending DNA samples across the street to the Joslin lab for automated sequencing.
After creating and cloning tailspike genes with the intended buried core mutations, we then characterized their folding and aggregation properties both in vivo in E. coli and in vitro. The denaturing gel below shows pellet and supernatant fractions of E. coli cells expressing the indicated tailspike chain. Replacement of the bulky aromatic side chains with increasingly smaller side chains (Leu > Val > Ala) completely blocked the formation of thermostable tailspike trimers under normal culture conditions. Instead, tailspike chains accumulated in the insoluble pellet fraction – as photogenic inclusion bodies. The most severe buried core mutants with Ala replacing Phe could not be rescued when produced or refolded even at the lowest possible temperatures. Based on these results, we concluded that these stacked side chains and others were critical to the correct folding of the parallel beta-helix domain.
Looking back, my brief time as a postdoc under Jon’s supervision was very productive, at least in terms of publications. Jon and I are co-authors on eight articles and chapters and one news and views commentary, not bad for a 3.5- year collaboration. I do remember spending the better part of two days sitting next to Jonathan in front of a big Power Mac monitor painstakingly reviewing word by word, and revising sentence by sentence, my first research manuscript in his lab, and wondering when he would tire. He didn’t. He persisted as always. Even our proofs were marked up quite extensively. I remember another lesson about science communication very clearly. In a draft abstract for a conference submission, I had started a sentence with the words “I am interested in…” Jonathan crossed out these words and explained that the conference attendees could care less what you the presenter are interested in. They want to know if you have learned anything that can help them advance their research projects. Full stop. I have lost count how many times since then that I have deleted these same words from early drafts of abstracts and manuscripts written by colleagues and interns.
On a personal note, Jonathan was from my experience ahead of the times in terms of work-life balance. My then-wife, Sharon Demorest, and I had two young children at home, as did Jon and Jackie. At one point, to supplement my meager postdoc income, I took on a Saturday only job as a “distribution generalist” for the Boston Globe, helping carriers pick up their stacks of Sunday newspapers. Eventually I mentioned my moonlighting to Jon and he brushed it off, noting that he hadn’t noticed any decrease in my dependability and productivity in the lab and not to worry. I was so thankful for this support and reassurance. Finally, a highlight of my years in Cambridge (other than the time Steve Raso took me to the Halligan Club in Charlestown😉 to hang out with local firemen) was one night when Jon and Jackie hosted my parents and I for dinner at their home in Cambridge. My parents, visiting from Oregon, were deeply honored to be invited to the home of an MIT professor, but also a bit intimidated. Of course, they were totally charmed and put at ease by the hospitality and warmness of Jon and Jackie. I heard my mom retell that story with great fondness for years after.


REFERENCES
- Betts, S., C. Haase-Pettingell, K. Cook and J. King (2004). “Buried hydrophobic side-chains essential for the folding of the parallel beta-helix domains of the P22 tailspike.” Protein Sci 13(9): 2291-2303.
- Betts, S. and J. King (1999). “There’s a right way and a wrong way: in vivo and in vitro folding, misfolding and subunit assembly of the P22 tailspike.” Structure 7(6): R131-139.
- Betts, S. D. and J. King (1998). “Cold rescue of the thermolabile tailspike intermediate at the junction between productive folding and off-pathway aggregation.” Protein Sci 7(7): 1516-1523.

